Publications
Selected publications
*first authors #senior authors/corresponding authors
| Haas-Kogan DA, et al. Everolimus for Children With Recurrent or Progressive Low-Grade Glioma: Results From the Phase II PNOC001 Trial. Journal of Clinical Oncology (2023). doi:10.1200/JCO.23.01838. PDF • PNOC001 clinical trial design • Best Poster Award, 28th Society for Neuro-Oncology Annual Meeting, Canada | 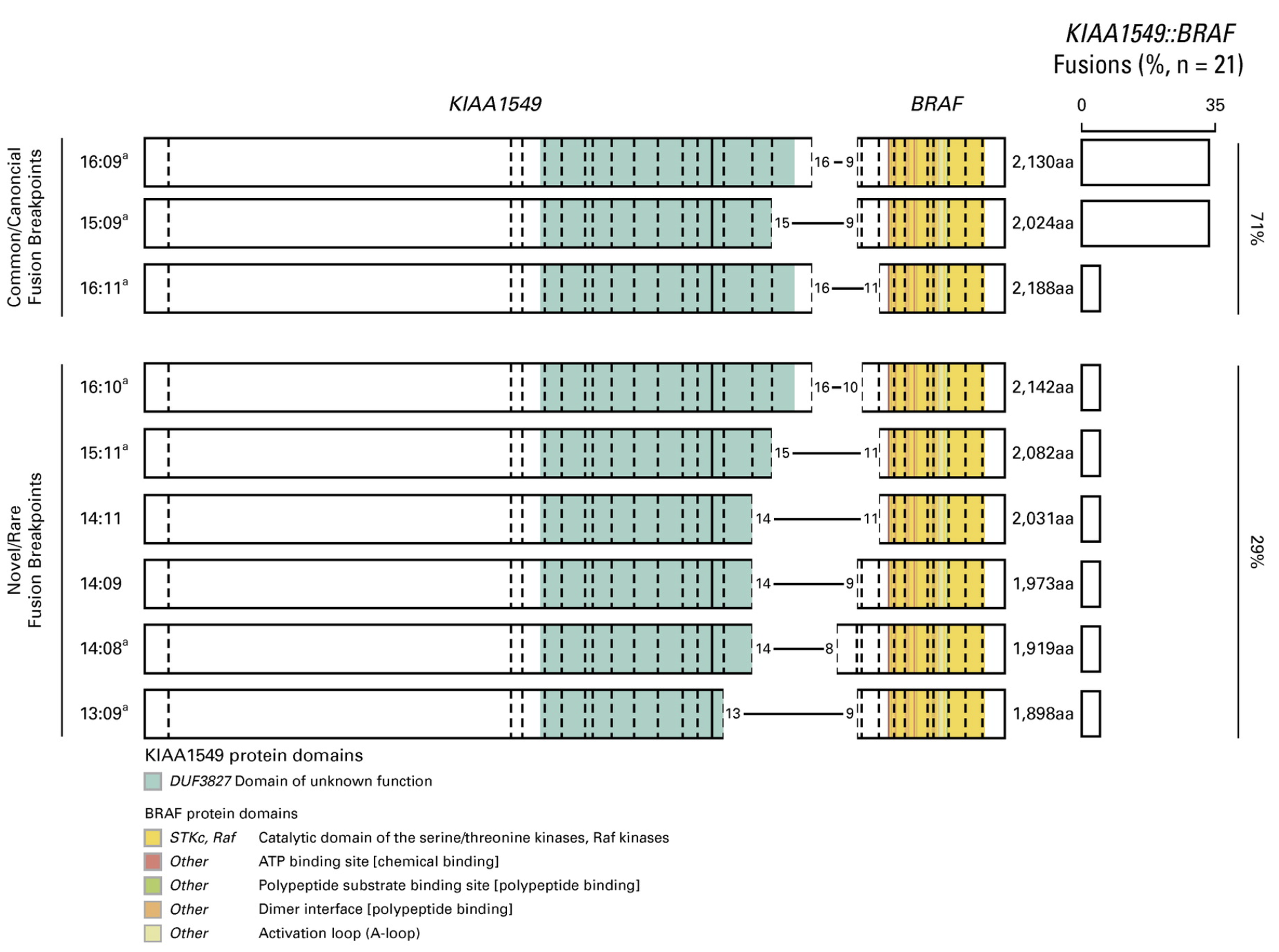 |
| Roberts HJ*, Ji S*, et al. Clinical, genomic, and epigenomic analyses of H3K27M-mutant diffuse midline glioma long-term survivors reveal a distinct group of tumors with MAPK pathway alterations. Acta Neuropathologica (2023). doi:10.1007/s00401-023-02640-7. PDF |  |
| Venneti S*, Kawakibi AR*, Ji S*, Waszak SM*, Sweha SR*, Mota M*, Pun M*, et al. Clinical efficacy of ONC201 in H3K27M-mutant diffuse midline gliomas is driven by disruption of integrated metabolic and epigenetic pathways. Cancer Discovery (2023). doi:10.1158/2159-8290.CD-23-0131. PDF |  |
| Waszak SM, Bourdeaut F, Nichols KE, Walsh MF. ELP1-related medulloblastoma predisposition syndrome. In: WHO Classification of Genetic Tumour Syndromes (2023). • Chapter is available online |  |
| Kline C*, Jain P*, Kilburn L*, et al. Upfront biology-guided therapy in diffuse intrinsic pontine glioma: therapeutic, molecular, and biomarker outcomes from PNOC003. Clinical Cancer Research (2022) doi:10.1158/1078-0432.CCR-22-0803. PDF • PNOC003 clinical trial design • PNOC003 genomic data portal |  |
| Pfister SM, Waszak SM. ELP1-medulloblastoma syndrome. In: WHO Classification of Central Nervous System Tumours (2021). • Chapter is available online and in print |  |
| Waszak SM*, Robinson GW*, et al. Germline Elongator mutations in Sonic Hedgehog medulloblastoma. Nature (2020) doi:10.1038/s41586-020-2164-5. PDF • Highlighted in Cancer Discovery and Neuro-Oncology • ELP1-MB was introduced as a new entity in OMIM, the WHO Classification of CNS Tumors, and the WHO Classification of Genetic Tumor Syndromes • PeCan ELP1-MB data portal • EMBL press release |  |
| ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium (Waszak SM*). Pan-cancer analysis of whole genomes. Nature (2020) doi:10.1038/s41586-020-1969-6. PDF • Manuscript is part of a special Nature issue • ICGC PCAWG data portal • EMBL research highlights and video interviews (video 1, video 2) • PCAWG Germline Working Group bioRxiv preprint (Waszak et al. 2017) |  |
| Begemann M*, Waszak SM*, et al. Germline GPR161 Mutations Predispose to Pediatric Medulloblastoma. Journal of Clinical Oncology (2019) doi:10.1200/jco.19.00577. PDF • Highlighted in Cancer Discovery • GPR161 was introduced as a novel genetic tumour syndrome in the WHO Classification of Genetic Tumour Syndromes • Updated clinical guideline: NCI | 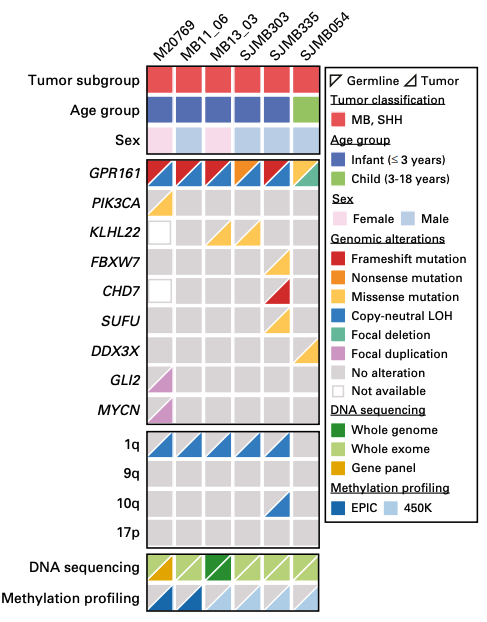 |
| Waszak SM*, Northcott PA*, et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. The Lancet Oncology (2018) doi:10.1016/S1470-2045(18)30242-0. PDF • Highlighted in Lancet Oncology, Cancer Discovery, ASCO Post • Updated clinical practise guidelines: NCCN, AWMF, EANO–EURACAN, NCI, eviQ • EMBL press release | 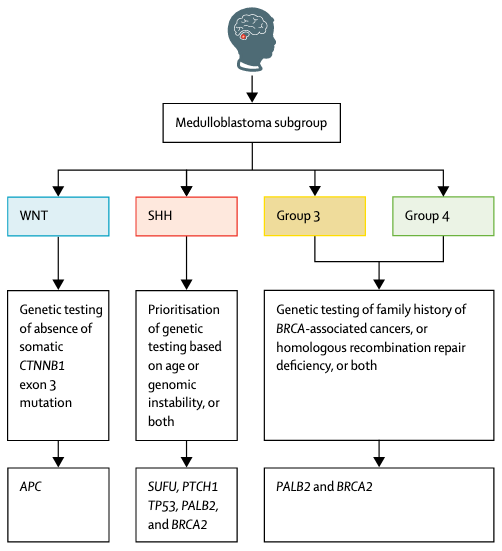 |
| Waszak SM*, Delaneau O*, et al. Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell (2015) doi: 10.1016/j.cell.2015.08.001. PDF • Highlighted in Nature Reviews Genetics and Cell • EPFL press release | 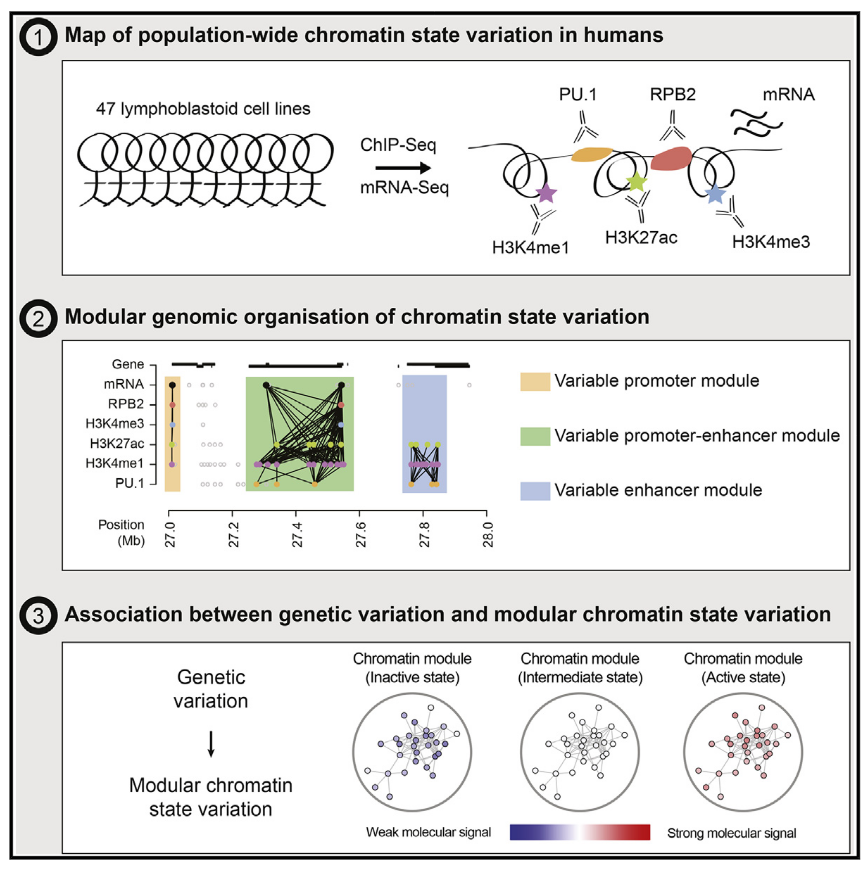 |
| Kilpinen H*, Waszak SM*, Gschwind AR*, et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science (2013) doi:10.1126/science.1242463. PDF • Highlighted in Nature Reviews Genetics and Science • EPFL press release |  |
Full list of publications
*first authors and #senior/corresponding authors
2023
84. Mueller S, Kline C, Franson A, van der Lugt J, Prados M, Waszak SM, Plasschaert S, Molinaro AM, Koschmann C, Nazarian J. Rational combination platform trial design for children and young adults with Diffuse Midline Glioma: a report from PNOC. Neuro-Oncology (2023). doi:10.1093/neuonc/noad181.
83. Haas-Kogan DA, Aboian MS, Minturn JE, Leary SES, Abdelbaki MS, Goldman S, Elster JD, Kraya A, Lueder MR, Ramakrishnan D, von Reppert M, Liu KX, Rokita JL, Resnick AC, Solomon DA, Phillips JJ, Prados M, Molinaro AM, Waszak SM#, Mueller S#. Everolimus for Children With Recurrent or Progressive Low-Grade Glioma: Results From the Phase II PNOC001 Trial. Journal of Clinical Oncology (2023). doi:10.1200/JCO.23.01838.
82. Roberts HJ*, Ji S*, Picca A, Sanson M, Garcia M, Snuderl M, Schüller U, Picart T, Ducray F, Green AL, Nakano Y, Sturm D, Abdullaev Z, Aldape K, Dang D, Kumar-Sinha C, Wu YM, Robinson D, Vo JN, Chinnaiyan AM, Cartaxo R, Upadhyaya SA, Mody R, Chiang J, Baker S, Solomon D, Venneti S, Pratt D, Waszak SM#, Koschmann C#. Clinical, genomic, and epigenomic analyses of H3K27M-mutant diffuse midline glioma long-term survivors reveal a distinct group of tumors with MAPK pathway alterations. Acta Neuropathologica (2023) doi:10.1007/s00401-023-02640-7.
81. Bonner ER, Dawood A, Gordish-Dressman H, Eze A, Bhattacharya S, Yadavilli S, Mueller S, Waszak SM#, Nazarian J#. Pan-cancer atlas of somatic core and linker histone mutations. npj Genomic Medicine (2023) Aug 28;8(1):1–14.
80. Venneti S*, Kawakibi AR*, Ji S*, Waszak SM*, Sweha SR*, Mota M*, Pun M*, Deogharkar A, Chung C, Tarapore RS, Ramage S, Chi A, Wen PY, Arrillaga-Romany I, Batchelor TT, Butowski NA, Sumrall A, Shonka N, Harrison RA, de Groot J, Mehta M, Hall MD, Daghistani D, Cloughesy TF, Ellingson BM, Beccaria K, Varlet P, Kim MM, Umemura Y, Garton H, Franson A, Schwartz J, Jain R, Kachman M, Baum H, Burant CF, Mottl SL, Cartaxo RT, John V, Messinger D, Qin T, Peterson E, Sajjakulnukit P, Ravi K, Waugh A, Walling D, Ding Y, Xia Z, Schwendeman A, Hawes D, Yang F, Judkins AR, Wahl D, Lyssiotis CA, de la Nava D, Alonso MM, Eze A, Spitzer J, Schmidt SV, Duchatel RJ, Dun MD, Cain JE, Jiang L, Stopka SA, Baquer G, Regan MS, Filbin MG, Agar NYR, Zhao L, Kumar-Sinha C, Mody R, Chinnaiyan A, Kurokawa R, Pratt D, Yadav VN, Grill J, Kline C, Mueller S, Resnick A, Nazarian J, Allen JE, Odia Y, Gardner SL, Koschmann C. Clinical efficacy of ONC201 in H3K27M-mutant diffuse midline gliomas is driven by disruption of integrated metabolic and epigenetic pathways. Cancer Discovery (2023). doi:10.1158/2159-8290.CD-23-0131.
79. Sanjaya P, Maljanen K, Katainen R, Waszak SM, Genomics England Research Consortium, Aaltonen LA, Stegle O, Korbel JO, Pitkänen E. Mutation-Attention (MuAt): deep representation learning of somatic mutations for tumour typing and subtyping. Genome Medicine (2023). doi:10.1186/s13073-023-01204-4.
78. Kolodziejczak AS, Guerrini-Rousseau L, Planchon JM, Ecker J, Selt F, Mynarek M, Obrecht D, Sill M, Autry RJ, Zhao E, Hirsch S, Amouyal E, Dufour C, Ayrault O, Torrejon J, Waszak SM, Ramaswamy V, Pentikainen V, Demir HA, Clifford SC, Schwalbe EC, Massimi L, Snuderl M, Galbraith K, Karajannis MA, Hill K, Li BK, Walsh M, White CL, Redmond S, Loizos L, Jakob M, Kordes UR, Schmid I, Hauer J, Blattmann C, Filippidou M, Piccolo G, Scheurlen W, Farrag A, Grund K, Sutter C, Pietsch T, Frank S, Schewe DM, Malkin D, Ben-Arush M, Sehested A, Wong TT, Wu KS, Liu YL, Carceller F, Mueller S, Stoller S, Taylor MD, Tabori U, Bouffet E, Kool M, Sahm F, von Deimling A, Korshunov A, von Hoff K, Kratz CP, Sturm D, Jones DTW, Rutkowski S, van Tilburg CM, Witt O, Bougeard G, Pajtler KW, Pfister SM, Bourdeaut F, Milde T. Clinical outcome of pediatric medulloblastoma patients with Li-Fraumeni syndrome. Neuro-Oncology (2023). doi:10.1093/neuonc/noad114.
77. Frioux C, Ansorge R, Özkurt E, Ghassemi Nedjad C, Fritscher J, Quince C, Waszak SM, Hildebrand F. Enterosignatures define common bacterial guilds in the human gut microbiome. Cell Host Microbe (2023). doi:10.1016/j.chom.2023.05.024.
76. Waszak SM, Bourdeaut F, Nichols KE, Walsh MF. ELP1-related medulloblastoma predisposition syndrome. In: Classification of Genetic Tumour Syndromes (1st ed.). World Health Organization (2023). https://tumourclassification.iarc.who.int/.
75. Jackson ER, Duchatel RJ, Staudt DE, Persson ML, Mannan A, Yadavilli S, Parackal S, Game S, Chong WC, Jayasekara WSN, Le Grand M, Kearney PS, Douglas AM, Findlay IJ, Germon ZP, McEwen HP, Beitaki TS, Patabendige A, Skerrett-Byrne DA, Nixon B, Smith ND, Day B, Manoharan N, Nagabushan S, Hansford JR, Govender D, McCowage GB, Firestein R, Howlett M, Endersby R, Gottardo NG, Alvaro F, Waszak SM, Larsen MR, Colino-Sanguino Y, Valdés-Mora F, Rakotomalala A, Meignan S, Pasquier E, Andre N, Hulleman E, Eisenstat DD, Vitanza NA, Nazarian J, Koschmann C, Mueller S, Cain JE, Dun MD. ONC201 in combination with paxalisib for the treatment of H3K27-altered diffuse midline glioma. Cancer Research (2023) doi:10.1158/0008-5472.CAN-23-0186.
74. Kratz CP, Smirnov D, Autry R, Jäger N, Waszak SM, Großhennig A, Berutti R, Wendorff M, Hainaut P, Pfister SM, Prokisch H, Ripperger T, Malkin D. Reply to Li and colleagues. Journal of the National Cancer Institute (2023). doi:10.1093/jnci/djad057.
73. Houlahan KE, Livingstone J, Fox NS, Kurganovs N, Zhu H, Sietsma Penington J, Jung CH, Yamaguchi TN, Heisler LE, Jovelin R, Costello AJ, Pope BJ, Kishan AU, Corcoran NM, Bristow RG, Waszak SM, Weischenfeldt J, He HH, Hung RJ, Hovens CM, Boutros PC. A polygenic two-hit hypothesis for prostate cancer. Journal of the National Cancer Institute (2023). doi:10.1093/jnci/djad001.
72. Kratz CP, Smirnov D, Autry R, Jäger N, Waszak SM, Großhennig A, Berutti R, Wendorff M, Hainaut P, Pfister SM, Prokisch H, Ripperger T, Malkin D. Reply to Evans and Woodward. Journal of the National Cancer Institute (2023). doi:10.1093/jnci/djac224.
2022
71. Houlahan KE, Yuan J, Schwarz T, Livingstone J, Fox NS, Jaratlerdsiri W, van Riet J, Taraszka K, Kurganovs N, Zhu H, Penington JS, Jung CH, Yamaguchi TN, Jiang J, Heisler LE, Jovelin R, Ramanand SG, Bell C, O’Connor E, Mutambirwa SBA, Seo JH, Costello AJ, Pomerantz MM, Pope BJ, Zaitlen N, Kishan AU, Corcoran NM, Bristow RG, Waszak SM, Bornman RMS, Gusev A, Lolkema MP, Weischenfeldt J, Hung RJ, He HH, Hayes VM, Pasaniuc B, Freedman ML, Hovens CM, Mani RS, Boutros PC. Germline determinants of the prostate tumor genome. bioRxiv (2022). doi:10.1101/2022.11.16.516773v1.full
70. Burkert M, Blanc E, Thiessen N, Weber C, Toedling J, Monti R, Dombrowe VM, de Biase MS, Kaufmann TL, Haase K, Waszak SM, Eggert A, Beule D, Schulte JH, Ohler U, Schwarz RF. Copy-number dosage regulates telomere maintenance and disease-associated pathways in neuroblastoma. bioRxiv (2022). doi:10.1101/2022.08.16.504100v1.full
69. Kratz CP, Smirnov D, Autry R, Jäger N, Waszak SM, Großhennig A, Berutti R, Wendorff M, Hainaut P, Pfister SM, Prokisch H, Ripperger T, Malkin D. Heterozygous BRCA1/2 and Mismatch Repair Gene Pathogenic Variants in Children and Adolescents with Cancer. Journal of the National Cancer Institute (2022). doi:10.1093/jnci/djac151.
68. Kline C*, Jain P*, Kilburn L*, Bonner ER, Gupta N, Crawford JR, Banerjee A, Packer RJ, Villanueva-Meyer J, Luks T, Zhang Y, Kambhampati M, Zhang J, Yadavilli S, Zhang B, Gaonkar KS, Rokita JL, Kraya A, Kuhn J, Liang W, Byron S, Berens M, Molinaro A, Prados M, Resnick A#, Waszak SM#, Nazarian J#, Mueller S#. Upfront biology-guided therapy in diffuse intrinsic pontine glioma: therapeutic, molecular, and biomarker outcomes from PNOC003. Clinical Cancer Research (2022). doi:10.1158/1078-0432.CCR-22-0803.
67. Guerrini-Rousseau L, Masliah-Planchon J, Waszak SM, Alhopuro P, Benusiglio PR, Bourdeaut F, Brecht IB, Del Baldo G, Dhanda SK, Garrè ML, Gidding CEM, Hirsch S, Hoarau P, Jorgensen M, Kratz C, Lafay-Cousin L, Mastronuzzi A, Pastorino L, Pfister SM, Schroeder C, Smith MJ, Vahteristo P, Vibert R, Vilain C, Waespe N, Winship IM, Evans DG, Brugieres L. Cancer risk and tumour spectrum in 172 patients with a germline SUFU pathogenic variation: a collaborative study of the SIOPE Host Genome Working Group. Journal of Medical Genetics (2022). doi:10.1136/jmedgenet-2021-108385.
66. Burns D, Anokian E, Saunders EJ, Bristow RG, Fraser M, Reimand J, Schlomm T, Sauter G, Brors B, Korbel J, Weischenfeldt J, Waszak SM, Corcoran NM, Jung CH, Pope BJ, Hovens CM, Cancel-Tassin G, Cussenot O, Loda M, Sander C, Hayes VM, Dalsgaard Sorensen K, Lu YJ, Hamdy FC, Foster CS, Gnanapragasam V, Butler A, Lynch AG, Massie CE; CR-UK/Prostate Cancer UK, ICGC, The PPCG, Woodcock DJ, Cooper CS, Wedge DC, Brewer DS, Kote-Jarai Z, Eeles RA. Rare Germline Variants Are Associated with Rapid Biochemical Recurrence After Radical Prostate Cancer Treatment: A Pan Prostate Cancer Group Study. European Urology (2022). doi:10.1016/j.eururo.2022.05.007.
65. Llimos G, Gardeux V, Koch U, Kribelbauer JF, Hafner A, Alpern D, Pezoldt J, Litovchenko M, Russeil J, Dainese R, Moia R, Mahmoud AM, Rossi D, Gaidano G, Plass C, Lutsik P, Gerhauser C, Waszak SM, Boettiger A, Radtke F, Deplancke B. A leukemia-protective germline variant mediates chromatin module formation via transcription factor nucleation. Nature Communications (2022). doi:10.1038/s41467-022-29625-6.
64. Sanjaya P, Waszak SM, Stegle O, Korbel JO, Pitkänen E. Mutation-Attention (MuAt): deep representation learning of somatic mutations for tumour typing and subtyping. bioRxiv(2022). doi:10.1101/2022.03.15.483816.
63. Przystal JM*, Cosentino CC*, Yadavilli S*, Zhang J, Laternser S, Bonner ER, Prasad R, Dawood AA, Lobeto N, Chong WC, Biery MC, Myers C, Olson JM, Panditharatna EA, Kritzer B, Mourabit S, Vitanza NA, Filbin MG, De Iuliis GN, Dun MD, Koschmann C, Cain J, Grotzer MA, Waszak SM, Mueller S, Nazarian J. Imipridones affect tumor bioenergetics and promote cell lineage differentiation in diffuse midline gliomas. Neuro-Oncology (2022). doi:10.1093/neuonc/noac041.
62. Pfister SM, Waszak SM. ELP1-medulloblastoma syndrome. In: Classification of Central Nervous System Tumours (5th ed.). World Health Organization (2022). https://publications.iarc.fr/601.
61. Findlay IJ, De Iuliis GN, Duchatel RJ, Jackson ER, Vitanza NA, Cain JE, Waszak SM, Dun MD. Pharmaco-proteogenomic profiling of pediatric diffuse midline glioma to inform future treatment strategies. Oncogene (2022). doi:10.1038/s41388-021-02102-y.
60. Messinger D, Harris MK, Cummings JR, Thomas C, Yang T, Sweha SR, Woo R, Siddaway R, Burkert M, Stallard S, Qin T, Mullan B, Siada R, Ravindran R, Niculcea M, Dowling A, Bradin J, Ginn KF, Gener MAH, Dorris K, Vitanza NA, Schmidt SV, Spitzer J, Jiang L, Filbin MG, Cao X, Castro MG, Lowenstein PR, Mody R, Chinnaiyan A, Desprez PY, McAllister S, Dun MD, Hawkins C, Waszak SM, Venneti S, Koschmann C, Yadav VN. Therapeutic targeting of prenatal pontine ID1 signaling in diffuse midline glioma. Neuro-Oncology (2022). doi:10.1093/neuonc/noac141.
2021
59. Hübschmann D*, Kleinheinz K*, Wagener R*, Bernhart SH*, López C*, Toprak UH, Sungalee S, Ishaque N, Kretzmer H, Kreuz M, Waszak SM, Paramasivam N, Ammerpohl O, Aukema SM, Beekman R, Bergmann AK, Bieg M, Binder H, Borkhardt A, Borst C, Brors B, Bruns P, de Santa Pau EC, Claviez A, Doose G, Haake A, Karsch D, Haas S, Hansmann M-L, Hoell JI, Hovestadt V, Huang B, Hummel M, Jäger-Schmidt C, Kerssemakers JNA, Korbel JO, Kube D, Lawerenz C, Lenze D, Martens JHA, Ott G, Radlwimmer B, Reisinger E, Richter J, Rico D, Rosenstiel P, Rosenwald A, Schillhabel M, Stilgenbauer S, Stadler PF, Martín-Subero JI, Szczepanowski M, Warsow G, Weniger MA, Zapatka M, Valencia A, Stunnenberg HG, Lichter P, Möller P, Loeffler M, Eils R, Klapper W, Hoffmann S, Trümper L, Küppers R, Schlesner M, Siebert R. Mutational mechanisms shaping the coding and noncoding genome of germinal center derived B-cell lymphomas. Leukemia (2021). doi:10.1038/s41375-021-01251-z.
58. Llamazares Prada M, Espinet E, Mijošek V, Schwartz U, Lutsik P, Tamas R, Richter M, Behrendt A, Pohl ST, Benz NP, Muley T, Warth A, Heußel CP, Winter H, Landry JJM, Herth FJ, Mertens TC, Karmouty-Quintana H, Koch I, Benes V, Korbel JO, Waszak SM, Trumpp A, Wyatt DM, Stahl HF, Plass C, Jurkowska RZ. Versatile workflow for cell-type resolved transcriptional and epigenetic profiles from cryopreserved human lung. JCI Insight (2021). doi:10.1172/jci.insight.140443.
57. Bonner ER, Waszak SM, Grotzer MA, Mueller S, Nazarian J. Mechanisms of imipridones in targeting mitochondrial metabolism in cancer cells. Neuro-Oncology (2021). doi:10.1093/neuonc/noaa283.
2020
56. Kawakibi AR, Tarapore R, Gardner S, Thomas C, Cartaxo R, Yadav V, Chi A, Kurz S, Wen P, Arrillaga I, Batchelor T, Butowski N, Sumrall A, Shonka N, Harrison R, Groot JD, Mehta M, Odia Y, Hall M, Daghistani D, Cloughesy T, Ellingson B, Kim M, Umemura Y, Garton H, Franson A, Robertson P, Schwartz J, Marini B, Pai M, Phoenix T, Ji S, Cantor E, Miklja Z, Mullan B, Bruzek A, Siada R, Cummings J, Stallard S, Wierzbicki K, Paul A, Wolfe I, Dun M, Cain J, Jiang L, Filbin M, Vats P, Kumar-Sinha C, Mody R, Chinnaiyan A, Pratt D, Venneti S, Lu G, Mueller S, Resnick A, Nazarian J, Waszak SM, Allen J, Koschmann C. Clinical efficacy and predictive biomarkers of ONC201 in H3 K27M-mutant diffuse midline glioma. Research Square (2020). doi:10.21203/rs.3.rs-69706/v1.
55. Jiao W, Atwal G, Polak P, Karlic R, Cuppen E; PCAWG Tumor Subtypes and Clinical Translation Working Group, Danyi A, de Ridder J, van Herpen C, Lolkema MP, Steeghs N, Getz G, Morris QD, Stein LD, PCAWG Consortium (Waszak SM). A deep learning system accurately classifies primary and metastatic cancers using passenger mutation patterns. Nature Communications (2020). doi:10.1038/s41467-019-13825-8.
54. Cmero M, Yuan K, Ong CS, Schröder J; PCAWG Evolution and Heterogeneity Working Group, Corcoran NM, Papenfuss T, Hovens CM, Markowetz F, Macintyre G, PCAWG Consortium ((Waszak SM). Inferring structural variant cancer cell fraction. Nature Communications (2020) doi:10.1038/s41467-020-14351-8.
53. Rubanova Y, Shi R, Harrigan CF, Li R, Wintersinger J, Sahin N, Deshwar AG, PCAWG Evolution and Heterogeneity Working Group, Morris QD, PCAWG Consortium (Waszak SM). Reconstructing evolutionary trajectories of mutation signature activities in cancer using TrackSig. Nature Communications (2020) doi:10.1038/s41467-020-14352-7.
52. Bhandari V, Li CH, Bristow RG, Boutros PC; PCAWG Consortium (Waszak SM). Divergent mutational processes distinguish hypoxic and normoxic tumours. Nature Communications (2020) doi: 10.1038/s41467-019-14052-x.
51. Sieverling L, Hong C, Koser SD, Ginsbach P, Kleinheinz K, Hutter B, Braun DM, Cortés-Ciriano I, Xi R, Kabbe R, Park PJ, Eils R, Schlesner M; PCAWG Structural Variation Working Group, Brors B, Rippe K, Jones DTW, Feuerbach L, PCAWG Consortium (Waszak SM). Genomic footprints of activated telomere maintenance mechanisms in cancer. Nature Communications (2020) doi:10.1038/s41467-019-13824-9.
50. Bailey MH, Meyerson WU, Dursi LJ, Wang L-B, Dong G, Liang W-W, Weerasinghe A, Li S, Kelso S, MC3 Working Group, PCAWG Novel Somatic Mutation Calling Methods Working Group, Saksena G, Ellrott K, Wendl MC, Wheeler DA, Getz G, Simpson JT, Gerstein MB, Ding L, PCAWG Consortium (Waszak SM). Retrospective evaluation of whole exome and genome mutation calls in 746 cancer samples. Nature Communications (2020). doi:10.1038/s41467-020-18151-y.
49. Li HC, Prokopec SD, Sun RX, Yousif F, Schmitz N, PCAWG Tumour Subtypes and Clinical Translation, Boutros PC, PCAWG Consortium (Waszak SM). Sex differences in oncogenic mutational processes. Nature Communications (2020). doi:10.1038/s41467-020-17359-2
48. Erarslan-Uysal* B, Kunz* JB, Rausch* T, Richter-Pechańska* P, van Belzen IA, Frismantas V, Bornhauser B, Ordoñez-Rueada D, Paulsen M, Benes V, Stanulla M, Schrappe M, Cario G, Escherich G, Bakharevich K, Kirschner-Schwabe R, Eckert C, Loukanov T, Gorenflo M, Waszak SM, Bourquin JP, Muckenthaler MU, Korbel JO, Kulozik AE. Chromatin accessibility landscape of pediatric T-lymphoblastic leukemia and human T-cell precursors. EMBO Molecular Medicine (2020) doi:10.15252/emmm.202012104
47. Drainas AP, Lambuta RA, Ivanova I, Sarropoulos I, Smith ML, Efthymiopoulos T, Raeder B, Stutz AM, Waszak SM, Mardin BR, Korbel JO. Genome wide screens identify CUL3 as a key regulator of persistent proliferation and genome instability in TP53 deficient cells. Cell Reports (2020) doi: 10.1016/j.celrep.2020.03.029.
46. Waszak SM*, Robinson GW*, Gudenas BL, Smith KS, Forget A, Hamilton KV, Indersie E, Buchhalter I, Jaeger N, Sharma T, Rausch T, Kool M, Sturm D, Jones DTW, Vasilyeva A, Tatevossian R, Neale G, Bowers D, Bendel A, Partap S, Chintagumpala M, Crawford J, Gottardo N, Smith A, Dufour C, Rutkowski S, Eggen T, Wesenberg F, Kjaerheim K, Feychting M, Lannering B, Schuz J, Johansen J, Andersen TV, Roosli M, Kuehni C, Grotzer M, Remke M, Pajtler K, Milde T, Witt O, Ryzhova M, Korshunov A, Orr BA Ellison DW, Brugieres L, Lichter P, Nichols KE, Gajjar A, Ayrault O, Korbel JO, Northcott PA, Pfister SM. Germline Elongator mutations in Sonic Hedgehog medulloblastoma. Nature (2020) doi:10.1038/s41586-020-2164-5.
45. ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium (Waszak SM*). Pan-cancer analysis of whole genomes. Nature (2020) doi:10.1038/s41586-020-1969-6. *co-first author and member of the manuscript writing committee.
44. Yakneen S, Waszak SM, PCAWG Technical Working Group, Gertz M, Korbel JO, PCAWG Consortium. Butler enables rapid cloud-based analysis of thousands of human genomes. Nature Biotechnology (2020) doi:10.1038/s41587-019-0360-3.
43. PCAWG Transcriptome Core Group, Calabrese C, Davidson NR, Demircioglu D, Fonseca NA, He Y, Kahles A, Lehmann KV, Liu F, Shiraishi Y, Soulette CM, Urban L, Greger L, Li S, Liu D, Perry MD, Xiang Q, Zhang F, Zhang J, Bailey P, Erkek S, Hoadley KA, Hou Y, Huska MR, Kilpinen H, Korbel JO, Marin MG, Markowski J, Nandi T, Pan-Hammarstrom Q, Pedamallu CS, Siebert R, Stark SG, Su H, Tan P, Waszak SM, Yung C, Zhu S, Awadalla P, Creighton CJ, Meyerson M, Ouellette BFF, Wu K, Yang H, PCAWG Transcriptome Working Group, Brazma A, Brooks AN, Goke J, Ratsch G, Schwarz RF, Stegle O, Zhang Z, PCAWG Consortium. Genomic basis for RNA alterations in cancer. Nature (2020) doi:10.1038/s41586-020-1970-0.
42. Li Y, Roberts ND, Wala JA, Shapira O, Schumacher SE, Kumar K, Khurana E, Waszak S, Korbel JO, Haber JE, Imielinski M; PCAWG Structural Variation Working Group, Weischenfeldt J, Beroukhim R, Campbell PJ, PCAWG Consortium. Patterns of structural variation in human cancer genomes. Nature (2020) doi:10.1038/s41586-019-1913-9.
41. Rodriguez-Martin B, Alvarez EG, Baez-Ortega A, Zamora J, Supek F, Demeulemeester J, Santamarina M, Ju YS, Temes J, Garcia-Souto D, Detering H, Li Y, Rodriguez-Castro J, Dueso-Barroso A, Bruzos AL, Dentro SC, Blanco MG, Contino G, Ardeljan D, Tojo M, Roberts ND, Zumalave S, Edwards PAW, Weischenfeldt J, Puiggros M, Chong Z, Chen K, Lee EA, Wala JA, Raine K, Butler A, Waszak SM, Navarro FCP, Schumacher SE, Monlong J, Maura F, Bolli N, Bourque G, Gerstein M, Park PJ, Wedge DC, Beroukhim R, Torrents D, Korbel JO, Martincorena I, Fitzgerald RC, Van Loo P, Kazazian HH, Burns KH, PCAWG Structural Variation Working Group, Campbell PJ, Tubio JMC, PCAWG Consortium. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nature Genetics (2020) doi:10.1038/s41588-019-0562-0.
40. Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao G, Hornshoj H, Hess JM, Juul RI, Lin Z, Feuerbach L, Sabarinathan R, Madsen T, Kim J, Mularoni L, Shuai S, Lanzos A, Herrmann C, Maruvka YE, Shen C, Amin SB, Bandopadhayay P, Bertl J, Boroevich KA, Busanovich J, Carlevaro-Fita J, Chakravarty D, Chan CWY, Craft D, Dhingra P, Diamanti K, Fonseca NA, Gonzalez-Perez A, Guo Q, Hamilton MP, Haradhvala NJ, Hong C, Isaev K, Johnson TA, Juul M, Kahles A, Kahraman A, Kim Y, Komorowski J, Kumar K, Kumar S, Lee D, Lehmann KV, Li Y, Liu EM, Lochovsky L, Park K, Pich O, Roberts ND, Saksena G, Schumacher SE, Sidiropoulos N, Sieverling L, Sinnott-Armstrong N, Stewart C, Tamborero D, Tubio JMC, Umer HM, Uuskula-Reimand L, Wadelius C, Wadi L, Yao X, Zhang CZ, Zhang J, Haber JE, Hobolth A, Imielinski M, Kellis M, Lawrence MS, von Mering C, Nakagawa H, Raphael BJ, Rubin MA, Sander C, Stein LD, Stuart JM, Tsunoda T, Wheeler DA, Johnson R, Reimand J, Gerstein M, Khurana E, Campbell PJ, Lopez-Bigas N, PCAWG Drivers and Functional Interpretation Working Group, PCAWG Structural Variation Working Group, Weischenfeldt J, Beroukhim R, Martincorena I, Pedersen JS, Getz G, PCAWG Consortium (Waszak SM). Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature (2020) doi:10.1038/s41586-019-1913-9.
39. Reyna MA, Haan D, Paczkowska M, Verbeke LPC, Vazquez M, Kahraman A, Pulido-Tamayo S, Barenboim J, Wadi L, Dhingra P, Shrestha R, Getz G, Lawrence MS, Pedersen JS, Rubin MA, Wheeler DA, Brunak S, Izarzugaza JMG, Khurana E, Marchal K, von Mering C, Sahinalp SC, Valencia A, PCAWG Drivers and Functional Interpretation Working Group, Reimand J, Stuart JM, Raphael BJ, PCAWG Consortium (Waszak SM). Pathway and network analysis of more than 2500 whole cancer genomes. Nature Communications (2020) doi:10.1038/s41467-020-14367-0.
38. Paczkowska M, Barenboim J, Sintupisut N, Fox NS, Zhu H, Abd-Rabbo D, Mee MW, Boutros PC, PCAWG Drivers and Functional Interpretation Working Group, Reimand J, PCAWG Consortium (Waszak SM). Integrative pathway enrichment analysis of multivariate omics data. Nature Communications (2020) doi:10.1038/s41467-019-13983-9.
37. Zhang Y, Chen F, Fonseca NA, He Y, Fujita M, Nakagawa H, Zhang Z, Brazma A, PCAWG Transcriptome Working Group, PCAWG Structural Variation Working Group, Creighton CJ, PCAWG Consortium (Waszak SM). High-coverage whole-genome analysis of 1220 cancers reveals hundreds of genes deregulated by rearrangement-mediated cis-regulatory alterations. Nature Communications (2020) doi:10.1038/s41467-019-13885-w.
36. Shuai S, PCAWG Drivers and Functional Interpretation Working Group, Gallinger S, Stein L, PCAWG Consortium (Waszak SM). Combined burden and functional impact tests for cancer driver discovery using DriverPower. Nature Communications (2020) doi:10.1038/s41467-019-13929-1.
35. Carlevaro-Fita J, Lanzos A, Feuerbach L, Hong C, Mas-Ponte D, Pedersen JS, PCAWG Drivers and Functional Interpretation Group, Johnson R, PCAWG Consortium (Waszak SM). Cancer LncRNA Census reveals evidence for deep functional conservation of long noncoding RNAs in tumorigenesis. Communications Biology (2020) doi:10.1038/s42003-019-0741-7.
2019
34. Begemann M*, Waszak SM*, Robinson GW, Jager N, Sharma T, Knopp C, Kraft F, Moser O, Mynarek M, Guerrini-Rousseau L, Brugieres L, Varlet P, Pietsch T, Bowers DC, Chintagumpala M, Sahm F, Korbel JO, Rutkowski S, Eggermann T, Gajjar A, Northcott PA, Elbracht M, Pfister SM, Kontny U, Kurth I. Germline GPR161 mutations predispose to pediatric medulloblastoma. Journal of Clinical Oncology (2019). doi:10.1200/jco.19.00577.
33. Ahad A, Stevanin M, Smita S, Mishra GP, Gupta D, Waszak S, Sarkar UA, Basak S, Gupta B, Acha-Orbea H, Raghav SK. NCoR1: putting the brakes on the dendritic cell immune tolerance. iScience (2019). doi:10.1016/j.isci.2019.08.024.
32. Lambo S, Gröbner S, Rausch T, Waszak SM, Schmidt C, Gorthi A, Romero C, Mauermann M, Brabetz S, Krausert S, Buchhalter I, Koster J, Zwijnenburg DA, Sill M, Hubner J-M, Mack N, Schwalm B, Ryzhova M, Hovestadt V, Papillon-Cavanagh S, Chan JA, Landgraf P, Ho B, Milde T, Witt O, Ecker J, Sahm F, Sumerauer D, Ellison DW, Orr BA, Darabi A, Haberler C, Figarella-Branger D, Wesseling P, Schittenhelm J, Remke M, Taylor MD, Gilda-Costa MJ, Lastowska M, Grajkowska W, Hasselblatt M, Hauser P, Pietsch T, Uro-Coste E, Bourdeaut F, Masliah-Planchon J, Rigau V, Alexandrescu S, Wolf S, Li X-N, Schuller U, Snuderl M, Karajannis MA, Giangaspero F, Jabado N, von Deimling A, Jones DTW, Korbel JO, von Hoff K, Lichter P, Huang A, Bishop A, Pfister SM, Korshunov A, Kool M. The molecular landscape of ETMR at diagnosis and relapse. Nature (2019) doi:10.1038/s41586-019-1815-x
31. Lopez C, Kleinheinz K, Aukema SM, Rohde M, Bernhart SH, Hubschmann D, Wagener R, Toprak UH, Raimondi F, Kreuz M, Waszak SM, Huang Z, Sieverling L, Paramasivam N, Seufert J, Sungalee S, Russell RB, Bausinger J, Kretzmer H, Ammerpohl O, Bergmann AK, Binder H, Borkhardt A, Brors B, Claviez A, Doose G, Feuerbach L, Haake A, Hansmann ML, Hoell J, Hummel M, Korbel JO, Lawerenz C, Lenze D, Radlwimmer B, Richter J, Rosenstiel P, Rosenwald A, Schilhabel MB, Stein H, Stilgenbauer S, Stadler PF, Szczepanowski M, Weniger MA, Zapatka M, Eils R, Lichter P, Loeffler M, Moller P, Trumper L, Klapper W, ICGC MMML-Seq Consortium, Hoffmann S, Kuppers R, Burkhardt B, Schlesner M, Siebert R. Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma. Nature Communications (2019). doi:10.1038/s41467-019-08578-3.
30. Hildebrand F, Moitinho-Silva L, Blasche S, Jahn MT, Gossmann TI, Huerta-Cepas J, Hercog R, Luetge M, Bahram M, Pryszlak A, Alves RJ, Waszak SM, Zhu A, Ye L, Costea PI, Aalvink S, Belzer C, Forslund SK, Sunagawa S, Hentschel U, Merten C, Patil KR, Benes V, Bork P. Antibiotics-induced monodominance of a novel gut bacterial order. Gut (2019). doi:10.1136/gutjnl-2018-317715.
2018
29. Waszak SM*, Northcott PA*, Buchhalter I, Robinson GW, Sutter C, Groebner S, Grund KB, Brugieres L, Jones DTW, Pajtler KW, Morrissy AS, Kool M, Sturm D, Chavez L, Ernst A, Brabetz S, Hain M, Zichner T, Segura-Wang M, Weischenfeldt J, Rausch T, Mardin BR, Zhou X, Baciu C, Lawerenz C, Chan JA, Varlet P, Guerrini-Rousseau L, Fults DW, Grajkowska W, Hauser P, Jabado N, Ra YS, Zitterbart K, Shringarpure SS, De La Vega FM, Bustamante CD, Ng HK, Perry A, MacDonald TJ, Hernaiz Driever P, Bendel AE, Bowers DC, McCowage G, Chintagumpala MM, Cohn R, Hassall T, Fleischhack G, Eggen T, Wesenberg F, Feychting M, Lannering B, Schuz J, Johansen C, Andersen TV, Roosli M, Kuehni CE, Grotzer M, Kjaerheim K, Monoranu CM, Archer TC, Duke E, Pomeroy SL, Shelagh R, Frank S, Sumerauer D, Scheurlen W, Ryzhova MV, Milde T, Kratz CP, Samuel D, Zhang J, Solomon DA, Marra M, Eils R, Bartram CR, von Hoff K, Rutkowski S, Ramaswamy V, Gilbertson RJ, Korshunov A, Taylor MD, Lichter P, Malkin D, Gajjar A, Korbel JO, Pfister SM. Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncology (2018). doi:10.1016/S1470-2045(18)30242-0.
28. Robinson GW,* Rudneva VA*, Buchhalter I, Billups CA, Waszak SM, Smith KS, Bowers DC, Bendel A, Fisher PG, Partap S, Crawford JR, Hassall T, Indelicato DJ, Boop F, Klimo P, Sabin ND, Patay Z, Merchant TE, Stewart CF, Orr BA, Korbel JO, Jones DTW, Sharma T, Lichter P, Kool M, Korshunov A, Pfister SM, Gilbertson RJ, Sanders RP, Onar-Thomas A, Ellison DW, Gajjar A, Northcott PA. Risk-adapted therapy for young children with medulloblastoma (SJYC07): therapeutic and molecular outcomes from a multicentre, phase 2 trial. Lancet Oncology (2018). doi:10.1016/S1470- 2045(18)30204-3.
27. Gerhauser C*, Favero F*, Risch T*, Simon R*, Feuerbach L*, Assenov Y*, Heckmann D*, Sidiropoulos N, Waszak SM, Hubschmann D, Urbanucci A, Girma EG, Kuryshev V, Klimczak LJ, Saini N, Stutz AM, Weichenhan D, Bottcher LM, Toth R, Hendriksen JD, Koop C, Lutsik P, Matzk S, Warnatz HJ, Amstislavskiy V, Feuerstein C, Raeder B, Bogatyrova O, Schmitz EM, Hube-Magg C, Kluth M, Huland H, Graefen M, Lawerenz C, Henry GH, Yamaguchi TN, Malewska A, Meiners J, Schilling D, Reisinger E, Eils R, Schlesner M, Strand DW, Bristow RG, Boutros PC, von Kalle C, Gordenin D, Sultmann H, Brors B, Sauter G, Plass C, Yaspo ML, Korbel JO, Schlomm T, Weischenfeldt J. Molecular evolution of early-onset prostate cancer identifies molecular risk markers and clinical trajectories. Cancer Cell (2018). doi:10.1016/j.ccell.2018.10.016.
26. Gröbner SN*, Worst BC*, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, Johann PD, Balasubramanian GP, Segura-Wang M, Brabetz S, Bender S, Hutter B, Sturm D, Pfaff E, Hubschmann D, Zipprich G, Heinold M, Eils J, Lawerenz C, Erkek S, Lambo S, Waszak S, Blattmann C, Borkhardt A, Kuhlen M, Eggert A, Fulda S, Gessler M, Wegert J, Kappler R, Baumhoer D, Burdach S, Kirschner-Schwabe R, Kontny U, Kulozik AE, Lohmann D, Hettmer S, Eckert C, Bielack S, Nathrath M, Niemeyer C, Richter GH, Schulte J, Siebert R, Westermann F, Molenaar JJ, Vassal G, Witt H, ICGC PedBrain-Seq Project, ICGC MMML-Seq Project, Burkhardt B, Kratz CP, Witt O, van Tilburg CM, Kramm CM, Fleischhack G, Dirksen U, Rutkowski S, Fruhwald M, von Hoff K, Wolf S, Klingebiel T, Koscielniak E, Landgraf P, Koster J, Resnick AC, Zhang J, Liu Y, Zhou X, Waanders AJ, Zwijnenburg DA, Raman P, Brors B, Weber UD, Northcott PA, Pajtler KW, Kool M, Piro RM, Korbel JO, Schlesner M, Eils R, Jones DTW, Lichter P, Chavez L, Zapatka M, Pfister SM. The landscape of genomic alterations across childhood cancers. Nature (2018). doi:10.1038/nature25480.
25. Richter-Pechanska P, Kunz JB, Bornhauser B, von Knebel Doeberitz C, Rausch T, Erarslan-Uysal B, Assenov Y, Frismantas V, Marovca B, Waszak SM, Zimmermann M, Seemann J, Happich M, Stanulla M, Schrappe M, Cario G, Escherich G, Bakharevich K, Kirschner-Schwabe R, Eckert C, Muckenthaler MU, Korbel JO, Bourquin JP, Kulozik AE. PDX models recapitulate the genetic and epigenetic landscape of pediatric T-cell leukemia. EMBO Molecular Medicine (2018). doi:10.15252/ emmm.201809443.
2017
24. Northcott PA*, Buchhalter I*, Morrissy AS*, Hovestadt V, Weischenfeldt J, Ehrenberger T, Gröbner S, Segura-Wang M, Zichner T, Rudneva VA, Warnatz HJ, Sidiropoulos N, Phillips AH, Schumacher S, Kleinheinz K, Waszak SM, Erkek S, Jones DTW, Worst BC, Kool M, Zapatka M, Jager N, Chavez L, Hutter B, Bieg M, Paramasivam N, Heinold M, Gu Z, Ishaque N, Jager-Schmidt C, Imbusch CD, Jugold A, Hubschmann D, Risch T, Amstislavskiy V, Gonzalez FGR, Weber UD, Wolf S, Robinson GW, Zhou X, Wu G, Finkelstein D, Liu Y, Cavalli FMG, Luu B, Ramaswamy V, Wu X, Koster J, Ryzhova M, Cho YJ, Pomeroy SL, Herold-Mende C, Schuhmann M, Ebinger M, Liau LM, Mora J, McLendon RE, Jabado N, Kumabe T, Chuah E, Ma Y, Moore RA, Mungall AJ, Mungall KL, Thiessen N, Tse K, Wong T, Jones SJM, Witt O, Milde T, Von Deimling A, Capper D, Korshunov A, Yaspo ML, Kriwacki R, Gajjar A, Zhang J, Beroukhim R, Fraenkel E, Korbel JO, Brors B, Schlesner M, Eils R, Marra MA, Pfister SM, Taylor MD, Lichter P. The whole-genome landscape of medulloblastoma subtypes. Nature (2017). doi:10.1038/nature22973.
23. Waszak SM*, Tiao G*, Zhu B*, Rausch T*, Muyas F, Rodriguez-Martin B, Rabionet R, Yakneen S, Escaramis G, Li Y, Saini N, Roberts SA, Demidov GM, Pitkanen E, Delaneau O, Heredia-Genestar JM, Weischenfeldt J, Shringarpure SS, Chen J, Nakagawa H, Alexandrov LB, Drechsel O, Dursi LJ, Segre AV, Garrison E, Erkek S, Habermann N, Urban L, Khurana E, Cafferkey A, Hayashi S, Imoto S, Aaltonen LA, Alvarez EG, Baez-Ortega A, Bailey M, Bosio M, Bruzos AL, Buchhalter I, Bustamante CD, Calabrese C, DiBiase A, Gerstein M, Holik AZ, Hua X, Huang K, Letunic I, Klimczak LJ, Koster R, Kumar S, McLellan M, Mashl J, Mirabello L, Newhouse S, Prasad A, Raetsch G, Schlesner M, Schwarz R, Sharma P, Shmaya T, Sidiropoulos N, Song L, Susak H, Tanskanen T, Tojo M, Wedge DC, Wright M, Wu Y, Ye K, Yellapantula VD, Zamora J, Butte AJ, Getz G, Simpson J, Ding L, Marques-Bonet T, Navarro A, Brazma A, Campbell P, Chanock SJ, Chatterjee N, Stegle O, Siebert R, Ossowski S, Harismendy O, Gordenin DA, Tubio JM, Vega FMDL, Easton DF, Estivill X, Korbel JO, on behalf of the ICGC/TCGA PCAWG Network. Germline determinants of the somatic mutation landscape in 2,642 cancer genomes. bioRxiv (2017). doi:10.1101/208330.
22. Sabarinathan R, Pich O, Martincorena I, Rubio-Perez C, Juul M, Wala J, Schumacher S, Shapira O, Sidiropoulos N, Waszak SM, Tamborero D, Mularoni L, Rheinbay E, Hornshoj H, Deu-Pons J, Muinos F, Bertl J, Guo Q, Creighton CJ, Weischenfeldt J, Korbel JO, Getz G, Campbell PJ, Pedersen JS, Beroukhim R, Gonzalez-Perez A, Lopez-Bigas N, on behalf of the ICGC/TCGA PCAWG Network. The whole-genome panorama of cancer drivers. bioRxiv (2017). doi:10.1101/190330.
2016
21. Weischenfeldt J*, Dubash T*, Drainas AP*, Mardin BR, Chen Y, Stutz AM, Waszak SM, Bosco G, Halvorsen AR, Raeder B, Efthymiopoulos T, Erkek S, Siegl C, Brenner H, Brustugun OT, Dieter SM, Northcott PA, Petersen I, Pfister SM, Schneider M, Solberg SK, Thunissen E, Weichert W, Zichner T, Thomas R, Peifer M, Helland A, Ball CR, Jechlinger M, Sotillo R, Glimm H, Korbel JO. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nature Genetics (2016). doi:10.1038/ng.3722.
20. Lin CY, Erkek S, Tong Y, Yin L, Federation AJ, Zapatka M, Haldipur P, Kawauchi D, Risch T, Warnatz HJ, Worst BC, Ju B, Orr BA, Zeid R, Polaski DR, Segura-Wang M, Waszak SM, Jones DT, Kool M, Hovestadt V, Buchhalter I, Sieber L, Johann P, Chavez L, Groschel S, Ryzhova M, Korshunov A, Chen W, Chizhikov VV, Millen KJ, Amstislavskiy V, Lehrach H, Yaspo ML, Eils R, Lichter P, Korbel JO, Pfister SM, Bradner JE, Northcott PA. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature (2016). doi:10.1038/nature16546.
19. Hutter S, Piro RM, Waszak SM, Kehrer-Sawatzki H, Friedrich RE, Lassaletta A, Witt O, Korbel JO, Lichter P, Schuhmann MU, Pfister SM, Tabori U, Mautner VF, Jones DTW. No correlation between NF1 mutation position and risk of optic pathway glioma in 77 unrelated NF1 patients. Human Genetics (2016). doi:10.1007/s00439-016-1646-x.
18. Korbel JO, Yakneen S, Waszak SM, Schlesner M, Eils R, Molnar-Gabor. A global initiative for cancer research: the pan-cancer analysis of whole genomes (PCAWG) project. Systembiologie (2016). i10.
17. Loviglio MN, Leleu M, Mannik K, Passeggeri M, Giannuzzi G, van der Werf I, Waszak SM, Zazhytska M, Roberts-Caldeira I, Gheldorf N, Migliavacca E, Alfaiz AA, Hippolyte L, Maillard AM, 2p15 Consortium, 16p11.2 Consortium, van Dijck A, Kooy RF, Salaville D, Rosenfeld JA, Shaffer LG, Andrieux J, Marshall C, Scherer SW, Shen Y, Gusella JF, Thorsteinsdottir U, Thorleifsson G, Dermitzakis ET, Deplancke B, Beckmann JS, Rougemont J, Jacquemont S, Reymond A. Chromosomal contacts connect loci associated with autism, BMI and head circumference phenotypes. Molecular Psychiatry (2016). doi:10.1038/mp.2016.84.
2015
16. Waszak SM*, Delaneau O*, Gschwind AR, Kilpinen H, Raghav SK, Witwicki RM, Orioli A, Wiederkehr M, Panousis NI, Yurovsky A, Romano-Palumbo L, Planchon A, Bielser D, Padioleau I, Udin G, Thurnheer S, Hacker D, Hernandez N, Reymond A, Deplancke B, Dermitzakis ET. Population variation and genetic control of modular chromatin architecture in humans. Cell (2015). doi: 10.1016/j.cell.2015.08.001.
15. Mardin BR, Drainas AP, Waszak SM, Weischenfeldt J, Isokane M, Stutz AM, Raeder B, Efthymiopoulos T, Buccitelli C, Segura-Wang M, Northcott P, Pfister SM, Lichter P, Ellenberg J, Korbel JO. A cell-based model system links chromothripsis with hyperploidy. Molecular Systems Biology (2015). doi:10.15252/msb.20156505.
2014
14. Waszak SM. A systems approach to elucidate the genetic architecture of molecular phenotypes. EPFL (2014). doi:10.5075/epfl-thesis-6179.
13. Waszak SM, Kilpinen H, Gschwind AR, Orioli A, Raghav SK, Witwicki RM, Migliavacca E, Yurovsky A, Lappalainen T, Hernandez N, Reymond A, Dermitzakis ET, Deplancke B. Identification and removal of low-complexity sites in allele-specific analysis of ChIP-seq data. Bioinformatics (2014). doi:10.1093/bioinformatics/btt667.
12. Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sorensen KD, Andersen LD, Andersen CL, Hulleman E, Wurdinger T, Ralfkiar E, Helin K, Gronbak K, Orntoft T, Waszak SM, Dahan O, Pedersen JS, Lund AH, Pilpel Y. A dual program for translation regulation in cellular proliferation and differentiation. Cell (2014). doi:10.1016/j.cell.2014.08.011.
11. Hammer C, Degenhardt F, Priebe L, Stutz AM, Heilmann S, Waszak SM, Schlattl A, Mangold E, Hoffmann P, Nothen MM, Rietschel M, Rappold G, Korbel J, Cichon S, Niesler B, MooDS Consortium. A common microdeletion affecting a hippocampus- and amygdala-specific isoform of tryptophan hydroxylase 2 is not associated with affective disorders. Bipolar Disorders (2014). doi:10.1111/bdi.12207.
10. Gubelmann C*, Schwalie PC*, Raghav SK*, Roder E, Delessa T, Kiehlmann E, Waszak SM, Corsinotti A, Udin G, Holcombe W, Rudofsky G, Trono D, Wolfrum C, Deplancke B. Identification of the transcription factor ZEB1 as a central component of the adipogenic gene regulatory network. eLife (2014). doi:10.7554/eLife.03346.
2013
9. Kilpinen H*, Waszak SM*, Gschwind AR*, Raghav SK, Witwicki RM, Orioli A, Migliavacca E, Wiederkehr M, Gutierrez-Arcelus M, Panousis NI, Yurovsky A, Lappalainen T, Romano-Palumbo L, Planchon A, Bielser D, Bryois J, Padioleau I, Udin G, Thurnheer S, Hacker D, Core LJ, Lis JT, Hernandez N, Reymond A, Deplancke B, Dermitzakis ET. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science (2013). doi:10.1126/science.1242463.
8. Waszak SM, Deplancke B. Rounding up natural gene expression variation during development. Developmental Cell (2013). doi:10.1016/j.devcel.2013.12.007.
7. Gubelmann C, Waszak SM, Isakova A, Holcombe W, Hens K, Iagovitina A, Feuz JD, Raghav SK, Simicevic J, Deplancke B. A yeast one-hybrid and microfluidics-based pipeline to map mammalian gene regulatory networks. Molecular Systems Biology (2013). doi:10.1038/msb.2013.38.
2012
6. Massouras A*, Waszak SM*, Albarca-Aguilera M, Hens K, Holcombe W, Ayroles JF, Dermitzakis ET, Stone EA, Jensen JD, Mackay TF, Deplancke B. Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS Genetics (2012). doi:10.1371/journal.pgen.1003055.
5. Raghav SK*, Waszak SM*, Krier I, Gubelmann C, Isakova A, Mikkelsen TS, Deplancke B. Integrative genomics identifies the corepressor SMRT as a gatekeeper of adipogenesis through the transcription factors C/EBPb and KAISO. Molecular Cell (2012). doi:10.1016/j.molcel.2012.03.017.
4. Olender T, Waszak SM, Viavant M, Khen M, Ben-Asher E, Reyes A, Nativ N, Wysocki CJ, Ge D, Lancet D. Personal receptor repertoires: olfaction as a model. BMC Genomics (2012). doi: 10.1186/1471-2164-13-414.
2011
3. Schlattl A, Anders S, Waszak SM, Huber W, Korbel JO. Relating CNVs to transcriptome data at fine resolution: assessment of the effect of variant size, type, and overlap with functional regions. Genome Research (2011). doi:10.1101/gr.122614.111.
2010
2. Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak SM, Habegger L, Rozowsky J, Shi M, Urban AE, Hong MY, Karczewski KJ, Huber W, Weissman SM, Gerstein MB, Korbel JO, Snyder M. Variation in transcription factor binding among humans. Science (2010). doi:10.1126/science.1183621.
1. Waszak SM, Hasin Y, Zichner T, Olender T, Keydar I, Khen M, Stutz AM, Schlattl A, Lancet D, Korbel JO. Systematic inference of copy-number genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity. PLoS Computational Biology (2010). doi:10.1371/journal.pcbi.1000988.